# There is no need to run these install lines below if you already have done so
install.packages("usethis")
usethis::use_course("https://github.com/andrewbtran/NICAR-2020-mapping/archive/master.zip")
# This section is in scripts/03_leaflet_choropleth.R
file.edit("scripts/03_leaflet_choropleth.R")leaflet choropleth
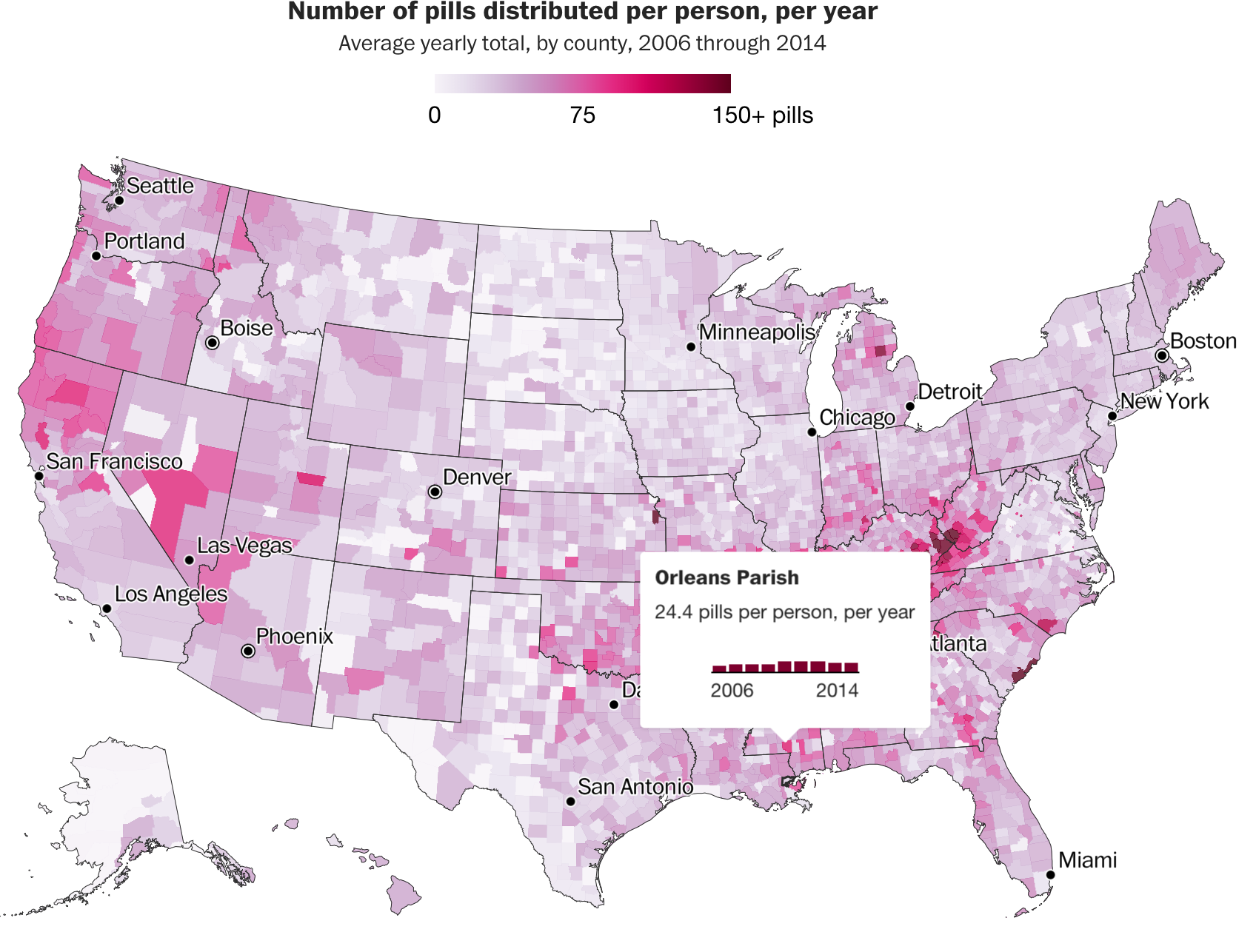
Choropleth or thematic maps are an effective and popular way to show geographic data.
The repo containing the data and scripts for this section is on Github. To follow along, simply run the lines of code below in R.
There’s a lot you must consider when making a choropleth map:
Adding interactivity to your graphic allows your most interested users to dig deep, explore the full data set, and establishes trust.
Making choropleths
To make a choropleth map, you first need a spatial data representing geographic boundaries. The term you hear most frequently are “shapefiles” but they could also be GeoJSONs, or TopoJSON, or KML, or even KMZ files, among others.
Basics
There are two underlying important pieces of information for spatial data:
- Coordinates of the object
- How the coordinates relate to a physical location on Earth
- Also known as coordinate reference system or CRS
CRS
- Geographic
- Uses three-dimensional model of the earth to define specific locations on the surface of the grid
- longitude (East/West) and latitude (North/South)
- Projected
- A translation of the three-dimensional grid onto a two-dimensional plane
Raster versus Vector data
Spatial data with a defined CRS can either be vector or raster data.
- Vector (Think Mapbox)
- Based on points that can be connected to form lines and polygons
- Located with in a coordinate reference system
- Example: Road map
- Raster (Think Google Maps and Leaflet)
- Are values within a grid system
- Example: Satellite imagery
sf vs sp
- An older package, sp, lets a user handle both vector and raster data.
- This class will focus on vector data and the sf package.
`It also takes much more effort to get your system ready for it (*shakes fist at gdal*). The main differences between the **sp** and **sf** packages are how they store CRS information. While **sp** uses spatial sub classes, **sf** stores data in data frames, allowing it to interact with **dplyr** methods we've learned so far. I encourage you to check out other spatial data analysis and modeling [classes](https://www.rspatial.org/) if you remain interested in this afterward.</aside>
Shape files
Though we refer to a shape file in the singular, it’s actually a collection of at least three basic files:
- .shp - lists shape and vertices
- .shx - has index with offsets
- .dbf - relationship file between geometry and attributes (data)
All files must be present in the directory and named the same (except for the file extension) to import correctly.
R can handle importing different kinds of file formats for spatial data, including KML and geojson and even topojson. We'll focus on shape files, which was created by ESRI in the '90s.The plan
- Map blank shape file we have already downloaded and is on the local machine
- Download opioid pain pill data for Louisiana via the arcos api
- Map data to blank shape file and map results
We can use magrittr %>% pipes because the leaflet package developers incorporated its function structure. We need three functions starting out:
leaflet()- initializes the leaflet functionaddTiles()- the underlying map tilesaddPolygons()- instead of dots, we’re adding Polygons, or shapes- Passing the argument
popupto the function with the variable NAME from the shape file
- Passing the argument
# This function checks if you don't have the correct packages installed yet
# If not, it will install it for you
packages <- c("tidyverse", "readr", "httpuv",
"leaflet", "sf", "tigris", "arcos",
"sp", "rmapshaper")
if (length(setdiff(packages, rownames(installed.packages()))) > 0) {
install.packages(setdiff(packages, rownames(installed.packages())), repos = "https://cran.us.r-project.org")
}
library(sf)
library(leaflet)
library(tidyverse)
library(arcos)
library(tigris)Importing shapefiles from the Census
There are more packages that people are producing in R that will pull polygons straight into R. For example, there’s an R package called Tigris that pulls from the Census TIGER shapefiles ftp server.
I’ve already downloaded the files to a folder called shapefiles. If you’ve downloaded the walkthrough files to your computer, they’re stored locally.
Importing shapefiles from your computer
# importing in the shapefile
counties <- st_read("shapefiles/us_counties.shp")Visualizing the shapefile
counties %>%
leaflet() %>%
addTiles() %>%
addPolygons(popup=~NAME)
This is how it looks raw. The Census shape files also include territories, like Guam and Puerto Rico.
When mapping, we’ll have to remember to exclude them if they show up.
Joining data to a shapefile
Let’s make a choropleth map based on number of opioids sold per state.
This data is pulled and transformed from the arcos api.
annual_summary <- read_csv("data/county_pill_summary.csv")
# If you'd like to build the data above from scratch
# using the arcos api, uncomment and run the lines below
#annual_counties <- summarized_county_annual(key="WaPo")
#annual_population <- county_population(key = "WaPo")
#annual <- left_join(annual_counties, annual_population)
#annual <- annual %>%
# mutate(pills_per_person=round(DOSAGE_UNIT/population,2))
#annual_summary <- annual %>%
# group_by(BUYER_COUNTY, BUYER_STATE, countyfips) %>%
# summarize(avg_pills_per_person=round(mean(pills_per_person, na.rm=T),2))
glimpse(annual_summary)Rows: 3,130
Columns: 4
$ BUYER_COUNTY <chr> "ABBEVILLE", "ACADIA", "ACCOMACK", "ADA", "ADAIR"…
$ BUYER_STATE <chr> "SC", "LA", "VA", "ID", "IA", "KY", "MO", "OK", "…
$ countyfips <chr> "45001", "22001", "51001", "16001", "19001", "210…
$ avg_pills_per_person <dbl> 18.73, 40.08, 20.87, 37.56, 25.62, 50.65, 49.02, …That’s some interesting looking data.
Tigris includes a useful function called geo_join() to bring together shape files and data frames. You may have used dplyr joins before, but geo_join() lets you join to non dataframe shape files.
We’ll also use a leaflet function called colorNumeric() that will turn the continuous variable of numbers of stores into a categorical variable by dividing it into bins and assigning it a hue of color we want. In this instance: Greens. Because, obviously.
Also setting up a separate array of pop up text for the map.
# Now we use the Tigris function geo_join to bring together
# the states shapefile and the sb_states dataframe -- GEOID and countyfips
# are the two columns they'll be joined by
counties_merged_annual <- geo_join(counties, annual_summary, "GEOID", "countyfips")
# Getting rid of rows with NA values
# Using the Base R method of filtering subset() because we're dealing with a SpatialPolygonsDataFrame and not a normal data frame, thus filter() wouldn't work
counties_merged_annual <- subset(counties_merged_annual, !is.na(avg_pills_per_person))
pal <- colorNumeric("Greens", domain=counties_merged_annual$avg_pills_per_person)
# Setting up the popup text
popup_sb <- paste0(counties_merged_annual$BUYER_COUNTY, ", ", counties_merged_annual$BUYER_STATE, "</br/> Average pills per person: \n", as.character(counties_merged_annual$avg_pills_per_person))Next, the map.
We’re adding a few more leaflet functions.
addProviderTiles()- instead ofaddTiles()- Uses the Leaflet Providers plugin to add different tiles to map
setView()- sets the starting position of the map- Centers it on defined coordinates with a specific zoom level
- Lots of arguments passed to
addPolygons()fillColor()fillOpacity()weightsmoothFactor()
addLegend()- same as in the previous section but with more customization
# Mapping it with the new tiles CartoDB.Positron
leaflet() %>%
addProviderTiles("CartoDB.Positron") %>%
setView(-98.483330, 38.712046, zoom = 4) %>%
addPolygons(data = counties_merged_annual ,
fillColor = ~pal(counties_merged_annual$avg_pills_per_person),
fillOpacity = 1,
weight = 0.9,
smoothFactor = 0.2,
stroke=TRUE,
color="white",
popup = ~popup_sb) %>%
addLegend(pal = pal,
values = counties_merged_annual$avg_pills_per_person,
position = "bottomright",
title = "Average pills per person")